import numpy as np
import matplotlib.pyplot as plt
import matplotlib.animation as animation
import matplotlib.gridspec as gridspec
np.random.seed(42)
# Generate data
num_points = 40
n_frames_per_angle = 2 # how many frames per angle. The lower - the faster.
X = np.random.randn(num_points, 2)
X = X @ np.array([[1.6, 0.0], [0.0, 0.4]])
# Normalize data
center_point = [0., 0.]
# PCA components
_, v = np.linalg.eig(X.T @ X)
v_main = v.T[0]
# Set up the grid
gs = gridspec.GridSpec(2, 1, height_ratios=[5, 1]) # Two rows, one column, with the first row 3 times the height of the second
fig = plt.figure(figsize=(5, 6)) # Adjust the total figure size as necessary
ax = plt.subplot(gs[0]) # The first subplot
ax2 = plt.subplot(gs[1]) # The second subplot
scatter = ax.scatter(X[:,0], X[:,1], color='b', label="Data")
direction_line, = ax.plot([], [], 'k')
ax.plot([-v_main[0]*3 + center_point[0],
v_main[0]*3 + center_point[0]],
[-v_main[1]*3 + center_point[1],
v_main[1]*3 + center_point[1]], label="First singular vector of X")
projection_points, = ax.plot([], [], 'ro', markersize=5, label="Projections")
projection_lines = [ax.plot([], [], 'r')[0] for _ in range(num_points)]
direction_line2, = ax2.plot([-3.5, 3.5], [0,0], 'k')
projections, = ax2.plot([],[], 'ro', markersize=7)
def init():
ax.axis('equal')
ax.grid(linestyle=":")
ax.scatter(x=center_point[0], y=center_point[1], c='k')
ax.legend(loc="upper right")
ax.set_title("PCA")
# ax.text(0.94, 0.945, "@fminxyz", transform=fig.transFigure,
# ha="right", va="top", fontsize=10, alpha=0.5)
ax2.set_xlim(-3.5, 3.5)
ax2.set_ylim(-1, 1)
w = np.array([0, 0])
ax2.grid(linestyle=":")
ax2.set_title("Projections on the First Principal Component\n"
f"Variance of the projections: {np.linalg.norm(X@w)**2:.1f}")
fig.tight_layout()
return scatter, direction_line, projection_points, projection_lines
def update(frame):
ax.set_xlim(-3.5+center_point[0], 3.5+center_point[0])
ax.set_ylim(-3.5+center_point[1], 3.5+center_point[1])
alpha = frame/n_frames_per_angle
w = np.array([np.cos(np.radians(alpha)), np.sin(np.radians(alpha))])
z = X @ w.reshape(-1, 1) @ w.reshape(1, -1) + center_point
for i in range(num_points):
projection_lines[i].set_data([X[i, 0], z[i, 0]], [X[i, 1], z[i, 1]])
projection_lines[i].set_color('r')
projection_points.set_data(z[:, 0], z[:, 1])
# distances = pdist(z)
# max_distance = np.max(distances)
# projection_points.set_label(f"Max Distance: {max_distance:.2f}")
direction_line.set_data([-w[0]*3 + center_point[0],
w[0]*3 + center_point[0]],
[-w[1]*3 + center_point[1],
w[1]*3 + center_point[1]])
ax2.set_xlim(-3.5, 3.5)
ax2.set_ylim(-1, 1)
projections.set_data(X@w, np.zeros(len(X@w)))
ax2.set_title("Projections on the First Principal Component\n"
f"Variance of the projections: {np.linalg.norm(X@w)**2:.1f}")
return direction_line, projection_points, projection_lines
ani = animation.FuncAnimation(fig, update,
frames=np.arange(0, n_frames_per_angle*180),
interval=1000/60, # 60 fps
init_func=init)
plt.close()
from IPython import display
html = display.HTML(ani.to_html5_video())
display.display(html)
# # Uncomment to save to the file
# ani.save("PCA_animation.mp4", writer='ffmpeg', fps=60, dpi=300)PCA intuition
Exercise: what’s wrong (1)?
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.animation as animation
import matplotlib.gridspec as gridspec
np.random.seed(42)
# Generate data
num_points = 40
n_frames_per_angle = 0.5 # how many frames per angle. The lower - the faster.
X = np.random.randn(num_points, 2)
X = X @ np.linalg.cholesky(np.array([[1, 0.6], [0.6, 0.6]]))
X = X - np.ones(2)
# Normalize data
center_point = [0., 0.]
# PCA components
_, v = np.linalg.eig(X.T @ X)
v_main = v.T[0]
# Set up the grid
gs = gridspec.GridSpec(2, 1, height_ratios=[5, 1]) # Two rows, one column, with the first row 3 times the height of the second
fig = plt.figure(figsize=(5, 6)) # Adjust the total figure size as necessary
ax = plt.subplot(gs[0]) # The first subplot
ax2 = plt.subplot(gs[1]) # The second subplot
scatter = ax.scatter(X[:,0], X[:,1], color='b', label="Data")
direction_line, = ax.plot([], [], 'k')
ax.plot([-v_main[0]*3 + center_point[0],
v_main[0]*3 + center_point[0]],
[-v_main[1]*3 + center_point[1],
v_main[1]*3 + center_point[1]], label="First singular vector of X")
projection_points, = ax.plot([], [], 'ro', markersize=5, label="Projections")
projection_lines = [ax.plot([], [], 'r')[0] for _ in range(num_points)]
direction_line2, = ax2.plot([-3.5, 3.5], [0,0], 'k')
projections, = ax2.plot([],[], 'ro', markersize=7)
def init():
ax.axis('equal')
ax.grid(linestyle=":")
ax.scatter(x=center_point[0], y=center_point[1], c='k')
ax.legend(loc="upper right")
ax.set_title("PCA")
# ax.text(0.94, 0.945, "@fminxyz", transform=fig.transFigure,
# ha="right", va="top", fontsize=10, alpha=0.5)
ax2.set_xlim(-3.5, 3.5)
ax2.set_ylim(-1, 1)
w = np.array([0, 0])
ax2.grid(linestyle=":")
ax2.set_title("Projections on the First Principal Component\n"
f"Variance of the projections: {np.linalg.norm(X@w)**2:.1f}")
fig.tight_layout()
return scatter, direction_line, projection_points, projection_lines
def update(frame):
ax.set_xlim(-3.5+center_point[0], 3.5+center_point[0])
ax.set_ylim(-3.5+center_point[1], 3.5+center_point[1])
alpha = frame/n_frames_per_angle
w = np.array([np.cos(np.radians(alpha)), np.sin(np.radians(alpha))])
z = X @ w.reshape(-1, 1) @ w.reshape(1, -1)
for i in range(num_points):
projection_lines[i].set_data([X[i, 0], z[i, 0]], [X[i, 1], z[i, 1]])
projection_lines[i].set_color('r')
projection_points.set_data(z[:, 0], z[:, 1])
# distances = pdist(z)
# max_distance = np.max(distances)
# projection_points.set_label(f"Max Distance: {max_distance:.2f}")
direction_line.set_data([-w[0]*3 + center_point[0],
w[0]*3 + center_point[0]],
[-w[1]*3 + center_point[1],
w[1]*3 + center_point[1]])
ax2.set_xlim(-3.5, 3.5)
ax2.set_ylim(-1, 1)
projections.set_data(X@w, np.zeros(len(X@w)))
ax2.set_title("Projections on the First Principal Component\n"
f"Variance of the projections: {np.linalg.norm(X@w)**2:.1f}")
return direction_line, projection_points, projection_lines
ani = animation.FuncAnimation(fig, update,
frames=np.arange(0, n_frames_per_angle*180),
interval=1000/60, # 60 fps
init_func=init)
plt.close()
from IPython import display
html = display.HTML(ani.to_html5_video())
display.display(html)
# # Uncomment to save to the file
# ani.save("PCA_animation.mp4", writer='ffmpeg', fps=60, dpi=300)Exercise: what’s wrong (2)?
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.animation as animation
import matplotlib.gridspec as gridspec
np.random.seed(42)
# Generate data
num_points = 30
n_frames_per_angle = 1 # how many frames per angle. The lower - the faster.
X = np.random.randn(num_points, 2)
# Normalize data
center_point = [0, 0]
# PCA components
eigs, v = np.linalg.eig(X.T @ X)
v_main = v.T[0]
# Set up the grid
gs = gridspec.GridSpec(2, 1, height_ratios=[5, 1]) # Two rows, one column, with the first row 3 times the height of the second
fig = plt.figure(figsize=(5, 6)) # Adjust the total figure size as necessary
ax = plt.subplot(gs[0]) # The first subplot
ax2 = plt.subplot(gs[1]) # The second subplot
scatter = ax.scatter(X[:,0], X[:,1], color='b', label="Data")
direction_line, = ax.plot([], [], 'k')
ax.plot([-v_main[0]*3 + center_point[0],
v_main[0]*3 + center_point[0]],
[-v_main[1]*3 + center_point[1],
v_main[1]*3 + center_point[1]], label="First singular vector of X")
projection_points, = ax.plot([], [], 'ro', markersize=5, label="Projections")
projection_lines = [ax.plot([], [], 'r')[0] for _ in range(num_points)]
direction_line2, = ax2.plot([-3.5, 3.5], [0,0], 'k')
projections, = ax2.plot([],[], 'ro', markersize=7)
def init():
ax.axis('equal')
ax.grid(linestyle=":")
ax.scatter(x=center_point[0], y=center_point[1], c='k')
ax.legend(loc="upper right")
ax.set_title("PCA")
# ax.text(0.94, 0.945, "@fminxyz", transform=fig.transFigure,
# ha="right", va="top", fontsize=10, alpha=0.5)
ax2.set_xlim(-3.5, 3.5)
ax2.set_ylim(-1, 1)
w = np.array([0, 0])
ax2.grid(linestyle=":")
ax2.set_title("Projections on the First Principal Component\n"
f"Variance of the projections: {np.linalg.norm(X@w)**2:.1f}")
fig.tight_layout()
return scatter, direction_line, projection_points, projection_lines
def update(frame):
ax.set_xlim(-3.5+center_point[0], 3.5+center_point[0])
ax.set_ylim(-3.5+center_point[1], 3.5+center_point[1])
alpha = frame/n_frames_per_angle
w = np.array([np.cos(np.radians(alpha)), np.sin(np.radians(alpha))])
z = X @ w.reshape(-1, 1) @ w.reshape(1, -1) + center_point
for i in range(num_points):
projection_lines[i].set_data([X[i, 0], z[i, 0]], [X[i, 1], z[i, 1]])
projection_lines[i].set_color('r')
projection_points.set_data(z[:, 0], z[:, 1])
# distances = pdist(z)
# max_distance = np.max(distances)
# projection_points.set_label(f"Max Distance: {max_distance:.2f}")
direction_line.set_data([-w[0]*3 + center_point[0],
w[0]*3 + center_point[0]],
[-w[1]*3 + center_point[1],
w[1]*3 + center_point[1]])
ax2.set_xlim(-3.5, 3.5)
ax2.set_ylim(-1, 1)
projections.set_data(X@w, np.zeros(len(X@w)))
ax2.set_title("Projections on the First Principal Component\n"
f"Variance of the projections: {np.linalg.norm(X@w)**2:.1f}")
return direction_line, projection_points, projection_lines
ani = animation.FuncAnimation(fig, update,
frames=np.arange(0, n_frames_per_angle*180),
interval=1000/60, # 60 fps
init_func=init)
plt.close()
from IPython import display
html = display.HTML(ani.to_html5_video())
display.display(html)
# # Uncomment to save to the file
# ani.save("PCA_animation.mp4", writer='ffmpeg', fps=60, dpi=300)Exercise: what’s “wrong” (3)?
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.animation as animation
import matplotlib.gridspec as gridspec
np.random.seed(42)
# Generate data
num_points = 60
n_frames_per_angle = 0.5 # how many frames per angle. The lower - the faster.
X_1 = np.random.randn(int(num_points/2), 2)
X_1 = X_1 @ np.array([[1.4, 0.0], [0.0, 0.2]])
X_2 = np.random.randn(int(num_points/2), 2)
X_2 = X_2 @ np.array([[1.4, 0.0], [0.0, 0.2]])
X_2 = X_2 + np.array([0, 2])
X = np.vstack([X_1, X_2])
# Normalize data
center_point = np.mean(X, axis=0)
X_std = X - center_point
# PCA components
_, v = np.linalg.eig(X_std.T @ X_std)
v_main = v.T[0]
# Set up the grid
gs = gridspec.GridSpec(2, 1, height_ratios=[5, 1]) # Two rows, one column, with the first row 3 times the height of the second
fig = plt.figure(figsize=(5, 6)) # Adjust the total figure size as necessary
ax = plt.subplot(gs[0]) # The first subplot
ax2 = plt.subplot(gs[1]) # The second subplot
scatter = ax.scatter(X[:,0], X[:,1], color='b', label="Data")
direction_line, = ax.plot([], [], 'k')
ax.plot([-v_main[0]*3 + center_point[0],
v_main[0]*3 + center_point[0]],
[-v_main[1]*3 + center_point[1],
v_main[1]*3 + center_point[1]], label="First eigenvector of X")
projection_points, = ax.plot([], [], 'ro', markersize=5, label="Projections")
projection_lines = [ax.plot([], [], 'r')[0] for _ in range(num_points)]
direction_line2, = ax2.plot([-3.5, 3.5], [0,0], 'k')
projections, = ax2.plot([],[], 'ro', markersize=7)
def init():
ax.axis('equal')
ax.grid(linestyle=":")
ax.scatter(x=center_point[0], y=center_point[1], c='k')
ax.legend(loc="upper right")
ax.set_title("PCA")
# ax.text(0.94, 0.945, "@fminxyz", transform=fig.transFigure,
# ha="right", va="top", fontsize=10, alpha=0.5)
ax2.set_xlim(-3.5, 3.5)
ax2.set_ylim(-1, 1)
w = np.array([0, 0])
ax2.grid(linestyle=":")
ax2.set_title("Projections on the First Principal Component\n"
f"Variance of the projections: {np.linalg.norm(X_std@w)**2:.1f}")
fig.tight_layout()
return scatter, direction_line, projection_points, projection_lines
def update(frame):
ax.set_xlim(-3.5+center_point[0], 3.5+center_point[0])
ax.set_ylim(-3.5+center_point[1], 3.5+center_point[1])
alpha = frame/n_frames_per_angle
w = np.array([np.cos(np.radians(alpha)), np.sin(np.radians(alpha))])
z = X_std @ w.reshape(-1, 1) @ w.reshape(1, -1) + center_point
for i in range(num_points):
projection_lines[i].set_data([X[i, 0], z[i, 0]], [X[i, 1], z[i, 1]])
projection_lines[i].set_color('r')
projection_points.set_data(z[:, 0], z[:, 1])
# distances = pdist(z)
# max_distance = np.max(distances)
# projection_points.set_label(f"Max Distance: {max_distance:.2f}")
direction_line.set_data([-w[0]*3 + center_point[0],
w[0]*3 + center_point[0]],
[-w[1]*3 + center_point[1],
w[1]*3 + center_point[1]])
ax2.set_xlim(-3.5, 3.5)
ax2.set_ylim(-1, 1)
projections.set_data(X_std@w, np.zeros(len(X_std@w)))
ax2.set_title("Projections on the First Principal Component\n"
f"Variance of the projections: {np.linalg.norm(X_std@w)**2:.1f}")
return direction_line, projection_points, projection_lines
ani = animation.FuncAnimation(fig, update,
frames=np.arange(0, n_frames_per_angle*180),
interval=1000/60, # 60 fps
init_func=init)
plt.close()
from IPython import display
html = display.HTML(ani.to_html5_video())
display.display(html)
# # Uncomment to save to the file
# ani.save("PCA_animation.mp4", writer='ffmpeg', fps=60, dpi=300)PCA with Iris
import numpy as np
import matplotlib.pyplot as plt
from sklearn.datasets import load_iris
from sklearn.preprocessing import StandardScaler
np.set_printoptions(suppress=True, precision=4)
dataset = load_iris()
A = dataset['data']
labels = dataset['target']
classes = dataset['target_names']
label_names = np.array([classes[label] for label in labels])
print('🤖: Dataset contains {} points in {}-dimensional space'.format(*A.shape))🤖: Dataset contains 150 points in 4-dimensional spaceprint('🌘 Mean value over each dimension before the normalization',np.mean(A, axis = 0))
# Data normalization with zero mean and unit variance
A_std = StandardScaler().fit_transform(A)
print('🌗 Mean value over each dimension after the normalization',np.mean(A_std, axis = 0))🌘 Mean value over each dimension before the normalization [5.8433 3.0573 3.758 1.1993]
🌗 Mean value over each dimension after the normalization [-0. -0. -0. -0.]# Main part
u,s,wh = np.linalg.svd(A_std)
print('🤖Shapes: \n A_std, {}\n u {}\n s {}. Singular values in descending order {} \n wh {}'.format(A_std.shape, u.shape, s.shape,s, wh.shape))🤖Shapes:
A_std, (150, 4)
u (150, 150)
s (4,). Singular values in descending order [20.9231 11.7092 4.6919 1.7627]
wh (4, 4)total_variance = sum(s)
variance_explained = [(i / total_variance)*100 for i in sorted(s, reverse=True)]
cumulative_variance_explained = np.cumsum(variance_explained)
xs = [0.5 + i for i in range(A_std.shape[1])]
plt.bar(xs, variance_explained, alpha=0.5, align='center',
label='Individual explained variance')
plt.step(xs, cumulative_variance_explained, where='mid',
label='Cumulative explained variance')
plt.ylabel('Explained variance')
plt.xlabel('Principal components')
plt.legend(loc='best')
plt.xticks(np.arange(A_std.shape[1]+1))
plt.show()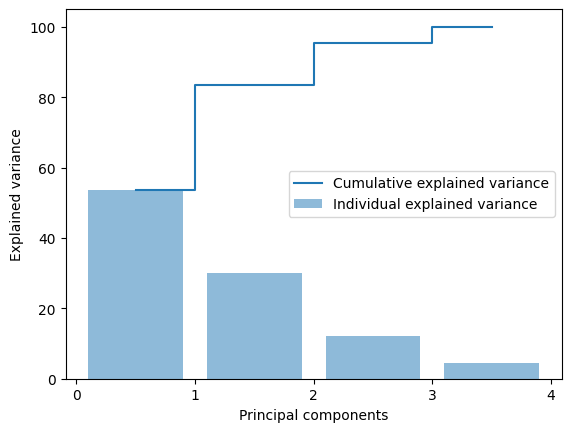
# Building projection matrix
rank = 2
w = wh.T
projections = u[:,:rank] @ np.diag(s[:rank])for label in classes:
plt.scatter(projections[label_names == label, 0],
projections[label_names == label, 1],
label = label)
plt.xlabel('PC 1')
plt.ylabel('PC 2')
plt.legend(loc='best')
plt.grid(linestyle=":")
plt.show()
# plt.savefig('pca_pr_iris.svg')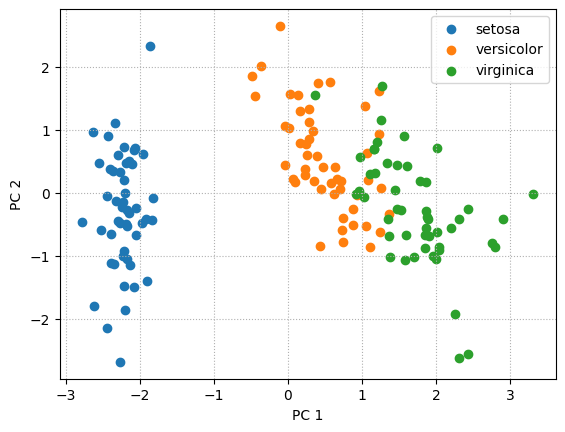
# Built-in approach
from sklearn.decomposition import PCA as sklearnPCA
sklearn_pca = sklearnPCA(n_components=2)
projections_sklearn = sklearn_pca.fit_transform(A_std)
for label in classes:
plt.scatter(projections_sklearn[label_names == label, 0],
projections_sklearn[label_names == label, 1],
label = label)
plt.xlabel('PC 1')
plt.ylabel('PC 2')
plt.legend(loc='best')
plt.grid(linestyle=":")
plt.show()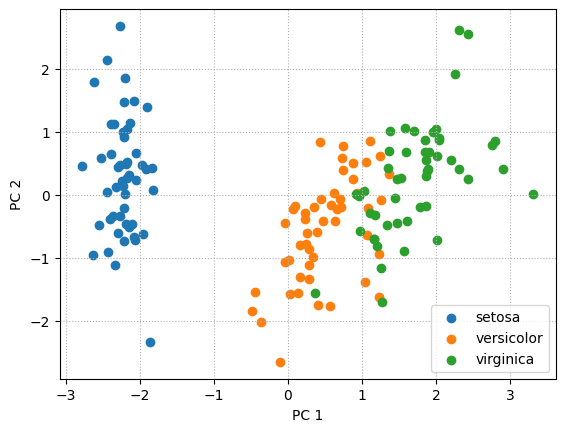
# Time comparison
def svd_projections(A_std):
u,s,wh = np.linalg.svd(A_std)
rank = 2
w = wh.T
return u[:,:rank] @ np.diag(s[:rank])
print('💎 SVD PCA running time')
%timeit svd_projections
print('💎 sklearn PCA running time')
%timeit sklearn_pca.fit_transformSVD PCA running time
11.4 ns ± 0.227 ns per loop (mean ± std. dev. of 7 runs, 100,000,000 loops each)
sklearn PCA running time
42.3 ns ± 0.484 ns per loop (mean ± std. dev. of 7 runs, 10,000,000 loops each)PCA with wine
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits import mplot3d
from sklearn.datasets import load_wine #lol
from sklearn.preprocessing import StandardScaler
dataset = load_wine()
A = dataset['data']
labels = dataset['target']
classes = dataset['target_names']
label_names = np.array([classes[label] for label in labels])
print('🤖: Dataset contains {} points in {}-dimensional space'.format(*A.shape))
# Data normalization with zero mean and unit variance
A_std = StandardScaler().fit_transform(A)
u,s,wh = np.linalg.svd(A_std)
total_variance = sum(s)
variance_explained = [(i / total_variance)*100 for i in sorted(s, reverse=True)]
cumulative_variance_explained = np.cumsum(variance_explained)
xs = [0.5 + i for i in range(A_std.shape[1])]
plt.bar(xs, variance_explained, alpha=0.5, align='center',
label='Individual explained variance')
plt.step(xs, cumulative_variance_explained, where='mid',
label='Cumulative explained variance')
plt.ylabel('Explained variance')
plt.xlabel('Principal components')
plt.legend(loc='best')
plt.xticks(np.arange(A_std.shape[1]+1))
plt.grid(linestyle=":")
plt.show()
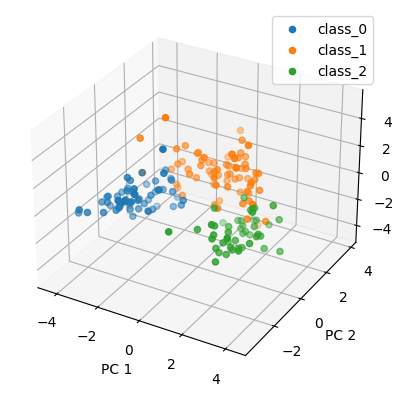
plt.figure()
rank = 3
projections = u[:,:rank] @ np.diag(s[:rank])
ax = plt.axes(projection="3d")
for label in classes:
ax.scatter3D(projections[label_names == label, 0],
projections[label_names == label, 1],
projections[label_names == label, 2],
label = label)
plt.xlabel('PC 1')
plt.ylabel('PC 2')
# plt.zlabel('PC 3')
plt.legend(loc='best')
plt.show()🤖: Dataset contains 178 points in 13-dimensional space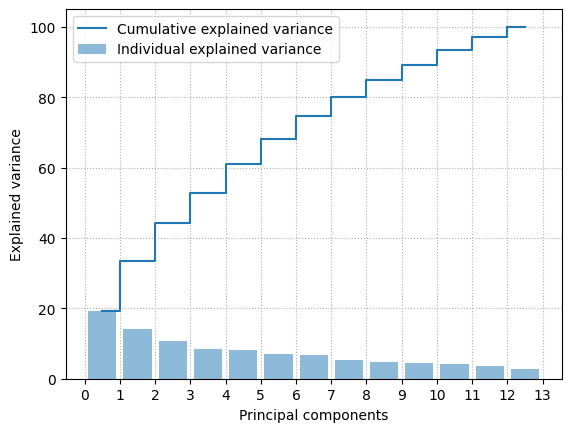
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits import mplot3d
import plotly.express as px
import pandas as pd
from sklearn.datasets import load_wine
from sklearn.preprocessing import StandardScaler
dataset = load_wine()
A = dataset['data']
labels = dataset['target']
classes = dataset['target_names']
label_names = np.array([classes[label] for label in labels])
print('🤖: Dataset contains {} points in {}-dimensional space'.format(*A.shape))
# Data normalization with zero mean and unit variance
A_std = StandardScaler().fit_transform(A)
u, s, wh = np.linalg.svd(A_std)
total_variance = sum(s)
variance_explained = [(i / total_variance)*100 for i in sorted(s, reverse=True)]
cumulative_variance_explained = np.cumsum(variance_explained)
xs = [0.5 + i for i in range(A_std.shape[1])]
plt.bar(xs, variance_explained, alpha=0.5, align='center',
label='Individual explained variance')
plt.step(xs, cumulative_variance_explained, where='mid',
label='Cumulative explained variance')
plt.ylabel('Explained variance')
plt.xlabel('Principal components')
plt.legend(loc='best')
plt.xticks(np.arange(A_std.shape[1]+1))
plt.grid(linestyle=":")
plt.show()
rank = 3
projections = u[:,:rank] @ np.diag(s[:rank])
df = pd.DataFrame(projections, columns=['PC1', 'PC2', 'PC3'])
df['label'] = label_names
fig = px.scatter_3d(df, x='PC1', y='PC2', z='PC3', color='label')
fig.update_traces(marker=dict(size=6),
selector=dict(mode='markers'))
fig.show()🤖: Dataset contains 178 points in 13-dimensional space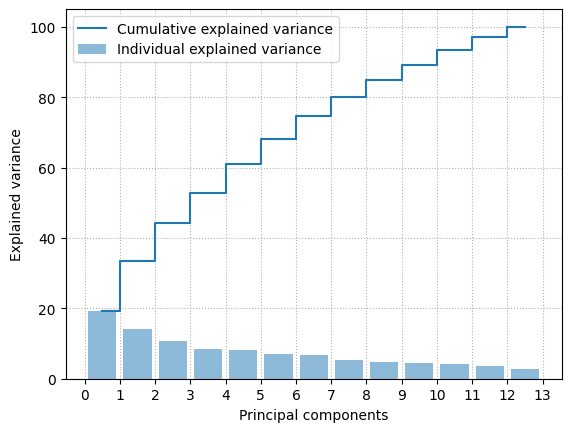
Unable to display output for mime type(s): application/vnd.plotly.v1+jsonMNIST PCA
import numpy as np
import plotly.express as px
import pandas as pd
import tensorflow as tf
# Load dataset
(train_images, train_labels), _ = tf.keras.datasets.mnist.load_data()
# Flatten images
train_images = train_images.reshape((train_images.shape[0], -1))
# Select 20 random images per class
selected_indices = []
for i in range(10):
indices = np.where(train_labels == i)[0]
selected_indices.extend(np.random.choice(indices, 1000, replace=False))
selected_images = train_images[selected_indices]
selected_labels = train_labels[selected_indices]
# Normalize the data
selected_images = selected_images / 255.0
# Apply PCA
from sklearn.decomposition import PCA
pca = PCA(n_components=2)
principal_components = pca.fit_transform(selected_images)
# Prepare data for plotting
pc_df = pd.DataFrame(data = principal_components, columns = ['PC1', 'PC2'])
pc_df['Label'] = selected_labels
pc_df['Label'] = pc_df['Label'].astype(str)
# Plot
fig = px.scatter(pc_df, x='PC1', y='PC2', color='Label',
color_discrete_sequence=px.colors.qualitative.Set1,
title='2D PCA of MNIST')
fig.show()Unable to display output for mime type(s): application/vnd.plotly.v1+jsonMNIST tSNE
import numpy as np
import plotly.express as px
import pandas as pd
import tensorflow as tf
from sklearn.manifold import TSNE
# Load dataset
(train_images, train_labels), _ = tf.keras.datasets.mnist.load_data()
# Flatten images
train_images = train_images.reshape((train_images.shape[0], -1))
# Select 20 random images per class
selected_indices = []
for i in range(10):
indices = np.where(train_labels == i)[0]
selected_indices.extend(np.random.choice(indices, 1000, replace=False))
selected_images = train_images[selected_indices]
selected_labels = train_labels[selected_indices]
# Normalize the data
selected_images = selected_images / 255.0
# Apply t-SNE
tsne = TSNE(n_components=2, random_state=0)
tsne_results = tsne.fit_transform(selected_images)
# Prepare data for plotting
tsne_df = pd.DataFrame(data = tsne_results, columns = ['Dim1', 'Dim2'])
tsne_df['Label'] = selected_labels
tsne_df['Label'] = tsne_df['Label'].astype(str)
# Plot
fig = px.scatter(tsne_df, x='Dim1', y='Dim2', color='Label',
color_discrete_sequence=px.colors.qualitative.Set1,
title='2D t-SNE of MNIST')
fig.show()Unable to display output for mime type(s): application/vnd.plotly.v1+jsonMNIST UMAP
!pip install umap-learnimport numpy as np
import plotly.express as px
import pandas as pd
import tensorflow as tf
import umap
# Load dataset
(train_images, train_labels), _ = tf.keras.datasets.mnist.load_data()
# Flatten images
train_images = train_images.reshape((train_images.shape[0], -1))
# Select 20 random images per class
selected_indices = []
for i in range(10):
indices = np.where(train_labels == i)[0]
selected_indices.extend(np.random.choice(indices, 1000, replace=False))
selected_images = train_images[selected_indices]
selected_labels = train_labels[selected_indices]
# Normalize the data
selected_images = selected_images / 255.0
# Apply UMAP
reducer = umap.UMAP(random_state=42)
embedding = reducer.fit_transform(selected_images)
# Prepare data for plotting
umap_df = pd.DataFrame(data = embedding, columns = ['Dim1', 'Dim2'])
umap_df['Label'] = selected_labels
umap_df['Label'] = umap_df['Label'].astype(str)
# Plot
fig = px.scatter(umap_df, x='Dim1', y='Dim2', color='Label',
color_discrete_sequence=px.colors.qualitative.Set1,
title='2D UMAP of MNIST')
fig.show()/Users/bratishka/.pyenv/versions/3.9.17/envs/benchmarx/lib/python3.9/site-packages/umap/umap_.py:1943: UserWarning:
n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
Unable to display output for mime type(s): application/vnd.plotly.v1+json